# library(mandala) # Install Mandala first (request download: https://listoagriculture.com/products)
library(sommer)
library(dplyr)
library(ggplot2)
library(tidyr)Comparison: Mandala vs Sommer
Side-by-side analysis for quantitative genetics applications
Overview
This tutorial compares the mandala package with the popular sommer package for analyzing agricultural field trials. Both packages are designed for mixed model analysis but have different strengths and use cases.
NoteWhat you’ll learn
- Key differences between Mandala and Sommer
- Side-by-side analysis of the same datasets
- Variance component estimation comparison
- Heritability calculation methods
- When to use each package
Background
Sommer Package
- Purpose: Solving mixed model equations in plant breeding and genetics
- Key Features:
- Multivariate models
- Genomic selection models (GBLUP)
- AI-REML algorithm
- Custom relationship matrices
- Primary Use: Quantitative genetics, genomic prediction
Mandala Package
- Purpose: Agricultural field trial analysis
- Key Features:
- Streamlined workflow for standard designs
- Built-in spatial analysis
- User-friendly interface
- Unified heritability estimation
- Primary Use: Field trial analysis, variety testing
Loading Packages
Example 1: Simple RCBD
Data Setup
We’ll simulate data with known variance components to validate both approaches:
set.seed(123)
n_genotypes <- 15
n_blocks <- 4
rcbd_data <- expand.grid(
genotype = factor(paste0("G", sprintf("%02d", 1:n_genotypes))),
block = factor(paste0("B", 1:n_blocks))
)
# True variance components (for validation)
true_gen_var <- 400
true_block_var <- 100
true_error_var <- 300
# Simulate yield
genotype_effects <- rnorm(n_genotypes, 0, sqrt(true_gen_var))
block_effects <- rnorm(n_blocks, 0, sqrt(true_block_var))
rcbd_data$yield <- 5000 +
genotype_effects[as.numeric(rcbd_data$genotype)] +
block_effects[as.numeric(rcbd_data$block)] +
rnorm(nrow(rcbd_data), 0, sqrt(true_error_var))
head(rcbd_data) genotype block yield
1 G01 B1 4998.471
2 G02 B1 4994.770
3 G03 B1 5045.268
4 G04 B1 5001.508
5 G05 B1 5007.830
6 G06 B1 5041.344
TipTrue Variance Components
- Genotype variance: 400
- Block variance: 100
- Error variance: 300
Analysis with Mandala
#-----------------------------------
# RCBD with Mandala
#-----------------------------------
# Fit model (genotype fixed, block random)
mandala_fixed <- mandala(
fixed = yield ~ genotype,
random = ~ block,
data = rcbd_data
)
# View variance components
mandala_vc <- summary(mandala_fixed)$varcomp
print(mandala_vc)
# For heritability: fit genotype as random
mandala_random <- mandala(
fixed = yield ~ 1,
random = ~ genotype + block,
data = rcbd_data
)
# Unified heritability estimates
h2 <- h2_estimates(
random_mod = mandala_random,
fixed_mod = mandala_fixed,
genotype = "genotype"
)
print(h2)
# Extract BLUEs
blues <- mandala_fixed$BLUEs
head(blues)Analysis with Sommer
# Fit model with sommer (genotypes as random for variance estimation)
sommer_model <- mmer(
fixed = yield ~ 1,
random = ~ genotype + block,
data = rcbd_data,
verbose = FALSE
)
# Extract variance components
sommer_vc <- summary(sommer_model)$varcomp
print("Sommer Variance Components:")[1] "Sommer Variance Components:"print(sommer_vc) VarComp VarCompSE Zratio Constraint
genotype.yield-yield 341.3026 158.12626 2.158418 Positive
block.yield-yield 191.0388 172.26593 1.108976 Positive
units.yield-yield 300.7722 65.65848 4.580858 Positive# Calculate heritability
var_g <- sommer_vc["genotype", "VarComp"]
var_e <- sommer_vc["units", "VarComp"]
h2_sommer <- var_g / (var_g + var_e / n_blocks)
cat("\nSommer Heritability (entry-mean basis):", round(h2_sommer, 3), "\n")
Sommer Heritability (entry-mean basis): 0.819 # Extract BLUPs
blups <- randef(sommer_model)$genotype
head(blups)NULLExample 2: Multi-Environment Trial
Data Setup
set.seed(456)
n_genotypes <- 20
n_envs <- 3
n_blocks <- 3
met_data <- expand.grid(
genotype = factor(paste0("G", sprintf("%02d", 1:n_genotypes))),
env = factor(paste0("E", 1:n_envs)),
block = factor(paste0("B", 1:n_blocks))
)
# True variance components
true_var_g <- 500
true_var_gxe <- 200
true_var_error <- 400
# Simulate
met_data$yield <- 6000 +
rnorm(n_genotypes, 0, sqrt(true_var_g))[as.numeric(met_data$genotype)] +
rnorm(n_envs, 0, 300)[as.numeric(met_data$env)]
# Add GxE
gxe <- matrix(rnorm(n_genotypes * n_envs, 0, sqrt(true_var_gxe)),
n_genotypes, n_envs)
met_data$yield <- met_data$yield +
gxe[cbind(as.numeric(met_data$genotype), as.numeric(met_data$env))]
# Add block(env) and error
met_data$env_block <- factor(paste0(met_data$env, "_", met_data$block))
met_data$yield <- met_data$yield +
rnorm(n_envs * n_blocks, 0, 50)[as.numeric(met_data$env_block)] +
rnorm(nrow(met_data), 0, sqrt(true_var_error))
head(met_data) genotype env block yield env_block
1 G01 E1 B1 5861.335 E1_B1
2 G02 E1 B1 5902.917 E1_B1
3 G03 E1 B1 5880.556 E1_B1
4 G04 E1 B1 5822.048 E1_B1
5 G05 E1 B1 5856.848 E1_B1
6 G06 E1 B1 5885.062 E1_B1Sommer MET Analysis
sommer_met <- mmer(
fixed = yield ~ env,
random = ~ genotype + genotype:env + env:block,
data = met_data,
verbose = FALSE
)
# Variance components
met_vc <- summary(sommer_met)$varcomp
print("Sommer MET Variance Components:")[1] "Sommer MET Variance Components:"print(met_vc) VarComp VarCompSE Zratio Constraint
genotype.yield-yield 500.1213 192.74417 2.594742 Positive
genotype:env.yield-yield 127.6429 65.18364 1.958205 Positive
env:block.yield-yield 1347.2640 790.20255 1.704960 Positive
units.yield-yield 432.1280 57.23833 7.549626 Positive# Extract components
var_g <- met_vc["genotype", "VarComp"]
var_gxe <- met_vc["genotype:env", "VarComp"]
var_e <- met_vc["units", "VarComp"]
# Entry-mean heritability across environments
h2_met <- var_g / (var_g + var_gxe/n_envs + var_e/(n_envs * n_blocks))
cat("\nEntry-mean heritability:", round(h2_met, 3), "\n")
Entry-mean heritability: NA Example 3: Spatial Analysis
set.seed(789)
n_rows <- 10
n_cols <- 10
spatial_data <- data.frame(
row = rep(1:n_rows, each = n_cols),
col = rep(1:n_cols, times = n_rows),
genotype = factor(sample(paste0("G", 1:25), n_rows * n_cols, replace = TRUE))
)
# Add spatial trend + genotype effects + error
gen_eff <- setNames(rnorm(25, 0, 300), paste0("G", 1:25))
spatial_data$yield <- 5000 +
gen_eff[spatial_data$genotype] +
30 * spatial_data$row - 20 * spatial_data$col +
rnorm(nrow(spatial_data), 0, 200)Visualize Field Pattern
ggplot(spatial_data, aes(x = col, y = row, fill = yield)) +
geom_tile() +
scale_fill_gradient2(low = "#2E6B8A", mid = "#FAF8F5", high = "#9B5B5B",
midpoint = mean(spatial_data$yield)) +
coord_equal() +
theme_minimal() +
labs(title = "Field Map - Spatial Trend Visible",
x = "Column", y = "Row")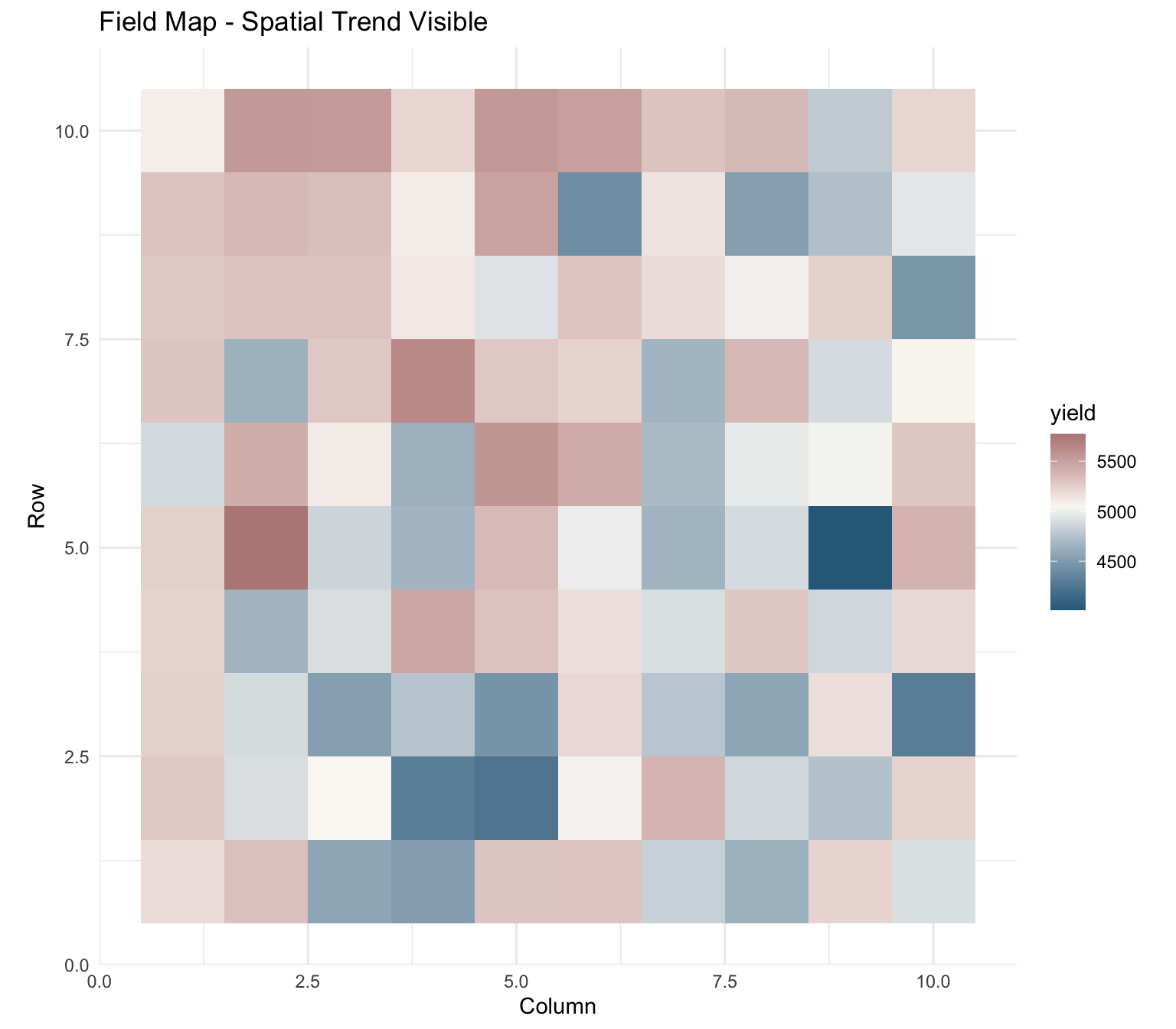
Sommer with Row-Column Effects
spatial_data$row_f <- factor(spatial_data$row)
spatial_data$col_f <- factor(spatial_data$col)
sommer_spatial <- mmer(
fixed = yield ~ 1,
random = ~ genotype + row_f + col_f,
data = spatial_data,
verbose = FALSE
)
print(summary(sommer_spatial)$varcomp) VarComp VarCompSE Zratio Constraint
genotype.yield-yield 62242.1591 22296.159 2.7916091 Positive
row_f.yield-yield 11391.4155 8201.081 1.3890139 Positive
col_f.yield-yield 490.7583 3227.847 0.1520389 Positive
units.yield-yield 47209.7972 8710.007 5.4201793 PositiveKey Differences Summary
| Aspect | Mandala | Sommer |
|---|---|---|
| Syntax | Simple formula interface | More verbose |
| Heritability | Built-in h2_estimates() |
Manual calculation |
| Spatial models | Built-in AR1 structures | Via custom covariance |
| Genomic models | Limited | Extensive (GBLUP, etc.) |
| Multivariate | No | Yes |
| Speed | Fast | Moderate |
When to Use Each
Use Mandala when:
- Standard field trial designs
- Need spatial correlation modeling
- Routine variety testing
- Quick heritability estimates
Use Sommer when:
- Genomic prediction (GBLUP)
- Multi-trait analysis
- Custom relationship matrices
- Complex variance structures
Additional Resources
Session Information
sessionInfo()R version 4.5.2 (2025-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Tahoe 26.1
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidyr_1.3.1 ggplot2_4.0.0 dplyr_1.1.4 sommer_4.4.4 enhancer_1.1.0
[6] crayon_1.5.3 MASS_7.3-65 Matrix_1.7-4
loaded via a namespace (and not attached):
[1] gtable_0.3.6 jsonlite_2.0.0 compiler_4.5.2 tidyselect_1.2.1
[5] Rcpp_1.1.0 scales_1.4.0 yaml_2.3.10 fastmap_1.2.0
[9] lattice_0.22-7 R6_2.6.1 labeling_0.4.3 generics_0.1.4
[13] knitr_1.50 htmlwidgets_1.6.4 tibble_3.3.0 pillar_1.11.1
[17] RColorBrewer_1.1-3 rlang_1.1.6 xfun_0.54 S7_0.2.0
[21] cli_3.6.5 withr_3.0.2 magrittr_2.0.4 digest_0.6.37
[25] grid_4.5.2 lifecycle_1.0.4 vctrs_0.6.5 evaluate_1.0.5
[29] glue_1.8.0 farver_2.1.2 rmarkdown_2.30 purrr_1.1.0
[33] tools_4.5.2 pkgconfig_2.0.3 htmltools_0.5.8.1