# library(mandala) # Install Mandala first (request download: https://listoagriculture.com/products)
library(dplyr)
library(ggplot2)
library(tidyr)Working with Example Datasets
Analyzing common experimental designs with Mandala
Overview
This tutorial demonstrates how to analyze real agricultural field trial datasets using the mandala package. We’ll work with common experimental designs and show how to properly specify models.
Loading Packages
Dataset 1: Randomized Complete Block Design (RCBD)
Data Description
A typical RCBD experiment with:
- 20 genotypes
- 3 replications (blocks)
- Response: grain yield (kg/ha)
# Create realistic RCBD dataset
set.seed(456)
n_genotypes <- 20
n_blocks <- 3
rcbd_data <- expand.grid(
genotype = factor(paste0("G", 1:n_genotypes)),
block = factor(paste0("B", 1:n_blocks))
)
# Simulate realistic yield data
rcbd_data$yield <- 5000
# Add genotype effects (some good, some poor yielders)
genotype_effects <- c(
rnorm(5, mean = 800, sd = 100), # High yielders
rnorm(10, mean = 0, sd = 150), # Average yielders
rnorm(5, mean = -600, sd = 100) # Low yielders
)
rcbd_data$yield <- rcbd_data$yield +
genotype_effects[as.numeric(rcbd_data$genotype)]
# Add block effects
block_effects <- c(200, -100, -50)
rcbd_data$yield <- rcbd_data$yield +
block_effects[as.numeric(rcbd_data$block)]
# Add error term
rcbd_data$yield <- rcbd_data$yield + rnorm(nrow(rcbd_data), mean = 0, sd = 300)
head(rcbd_data, 10) genotype block yield
1 G1 B1 5723.267
2 G2 B1 4881.472
3 G3 B1 5020.040
4 G4 B1 5046.350
5 G5 B1 4783.985
6 G6 B1 5113.979
7 G7 B1 4499.892
8 G8 B1 4729.488
9 G9 B1 5199.150
10 G10 B1 5735.364Exploratory Data Analysis
# Summary statistics by genotype
rcbd_summary <- rcbd_data %>%
group_by(genotype) %>%
summarise(
mean_yield = mean(yield),
sd_yield = sd(yield),
cv = (sd_yield / mean_yield) * 100
) %>%
arrange(desc(mean_yield))
head(rcbd_summary, 10)# A tibble: 10 × 4
genotype mean_yield sd_yield cv
<fct> <dbl> <dbl> <dbl>
1 G11 5760. 171. 2.97
2 G10 5734. 152. 2.65
3 G12 5589. 99.8 1.79
4 G13 5441. 349. 6.41
5 G1 5406. 413. 7.64
6 G16 5336. 389. 7.29
7 G20 5241. 87.5 1.67
8 G14 5232. 107. 2.05
9 G15 5197. 154. 2.96
10 G18 5159. 345. 6.68# Visualization
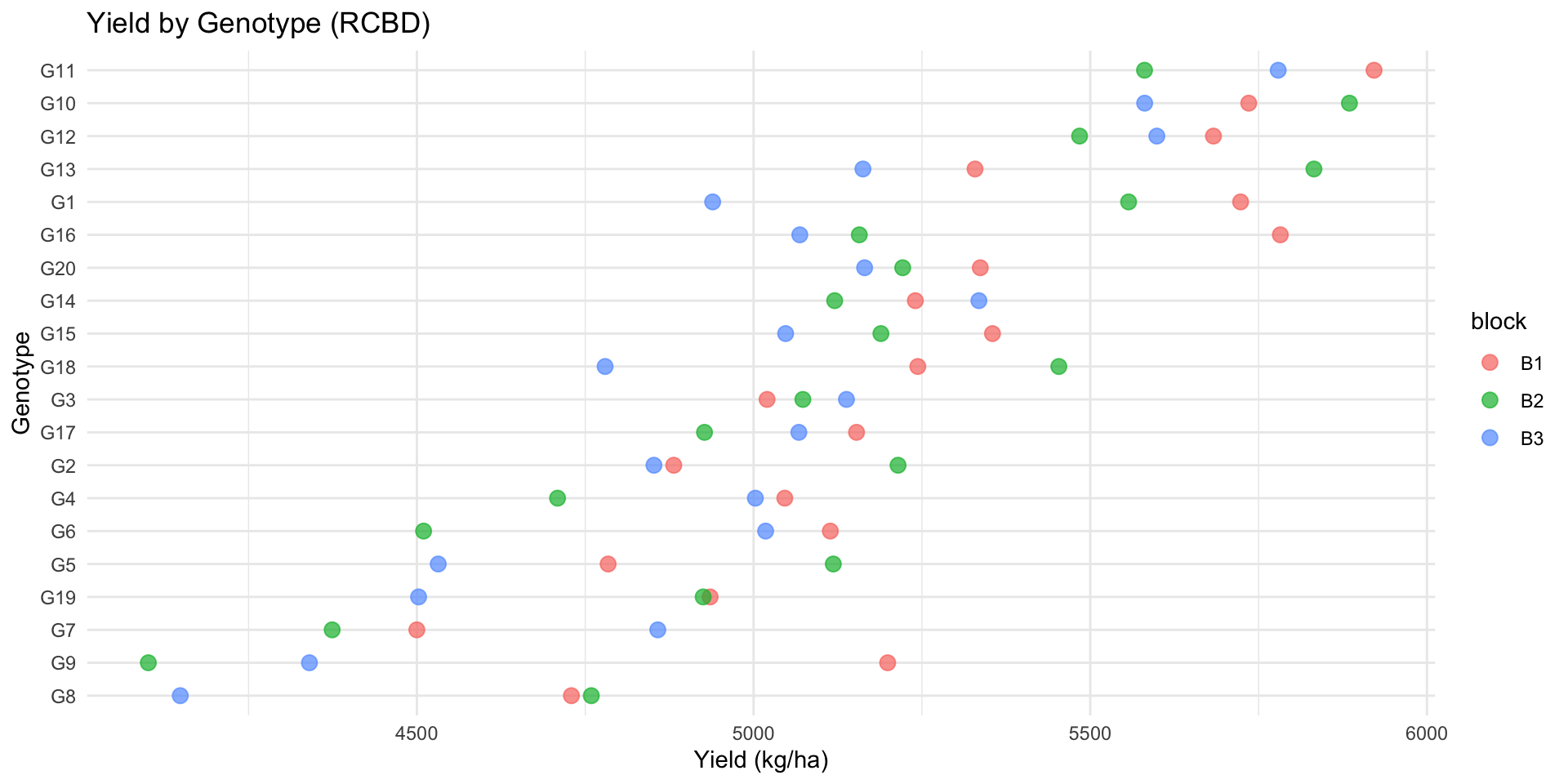
ggplot(rcbd_data, aes(x = reorder(genotype, yield), y = yield, color = block)) +
geom_point(size = 3, alpha = 0.7) +
coord_flip() +
theme_minimal() +
labs(
title = "Yield by Genotype (RCBD)",
x = "Genotype",
y = "Yield (kg/ha)"
)
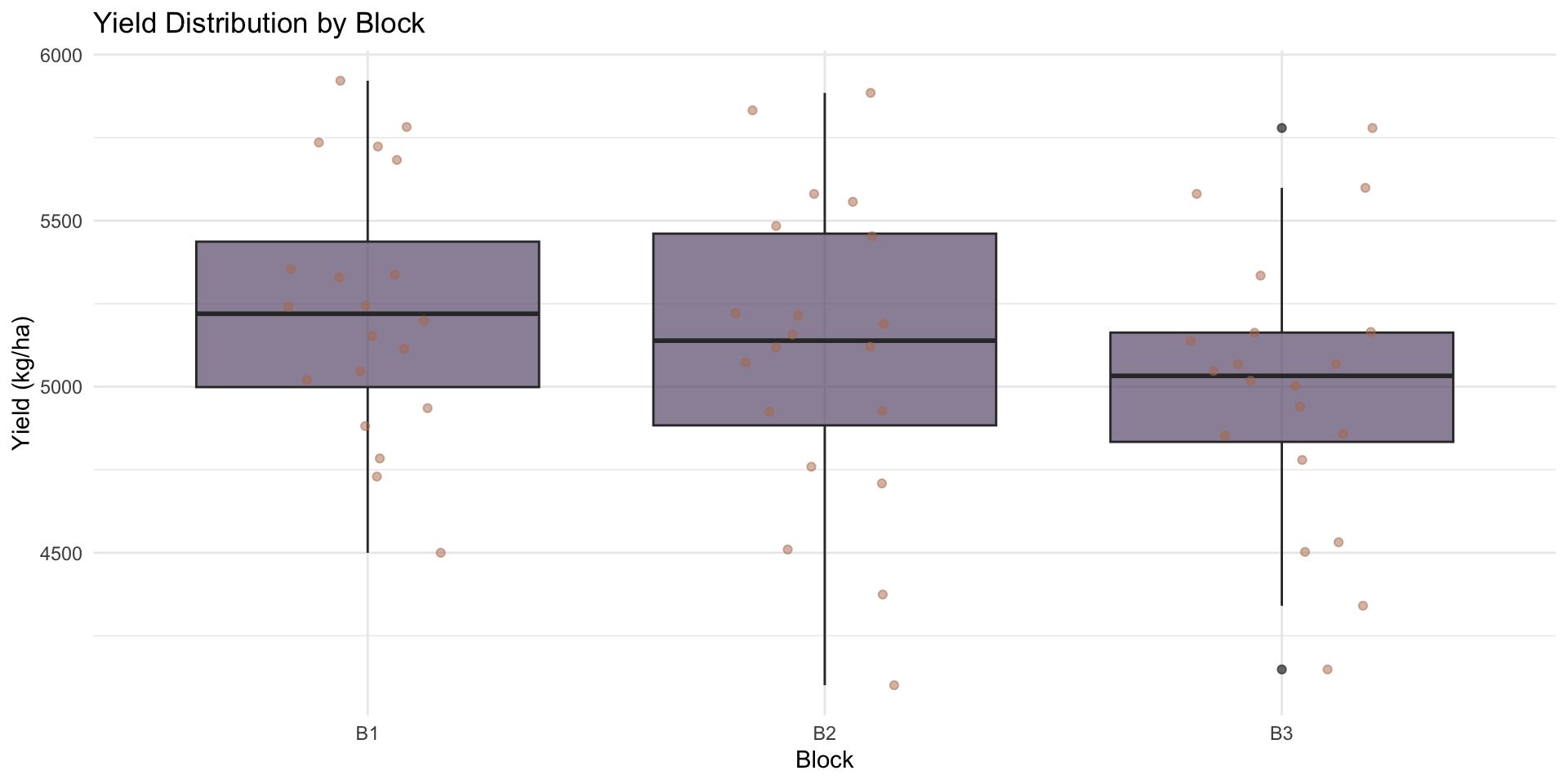
# Block effects
ggplot(rcbd_data, aes(x = block, y = yield)) +
geom_boxplot(fill = "#6B5A7D", alpha = 0.7) +
geom_jitter(width = 0.2, alpha = 0.5, color = "#B5795A") +
theme_minimal() +
labs(
title = "Yield Distribution by Block",
x = "Block",
y = "Yield (kg/ha)"
)
Model Fitting with Mandala
# Fit the RCBD model
rcbd_model <- mandala(
fixed = yield ~ genotype,
random = ~ block,
data = rcbd_data
)
# View model summary and variance components
summary(rcbd_model)
# Extract variance components table
var_components <- summary(rcbd_model)$varcomp
print(var_components)
# Heritability Estimation
rcbd_model_rand <- mandala(
fixed = yield ~ 1,
random = ~ genotype + block,
data = rcbd_data
)
h2_table <- h2_estimates(
random_mod = rcbd_model_rand,
fixed_mod = rcbd_model,
genotype = "genotype"
)
print(h2_table)
# Extract BLUEs and Rank Genotypes
blues <- rcbd_model$BLUEs
genotype_ranking <- blues %>% arrange(desc(estimate))
print(genotype_ranking)Dataset 2: Alpha-Lattice Design
Data Description
An alpha-lattice design with:
- 50 genotypes
- 2 replications
- Incomplete blocks of size 5 within each replication
set.seed(789)
n_genotypes <- 50
n_reps <- 2
block_size <- 5
n_blocks_per_rep <- n_genotypes / block_size
# Create design structure
alpha_data <- data.frame(
genotype = factor(rep(paste0("G", 1:n_genotypes), times = n_reps)),
rep = factor(rep(1:n_reps, each = n_genotypes))
)
# Assign incomplete blocks within reps
alpha_data$incomplete_block <- factor(
paste0(alpha_data$rep, ".",
rep(rep(1:n_blocks_per_rep, each = block_size), times = n_reps))
)
# Simulate yield data
alpha_data$yield <- 6000
# Add genotype effects
genotype_effects <- rnorm(n_genotypes, mean = 0, sd = 400)
alpha_data$yield <- alpha_data$yield +
genotype_effects[as.numeric(alpha_data$genotype)]
# Add rep effects
rep_effects <- c(300, -300)
alpha_data$yield <- alpha_data$yield +
rep_effects[as.numeric(alpha_data$rep)]
# Add incomplete block effects
n_incomplete_blocks <- n_reps * n_blocks_per_rep
incomplete_block_effects <- rnorm(n_incomplete_blocks, mean = 0, sd = 200)
alpha_data$yield <- alpha_data$yield +
incomplete_block_effects[as.numeric(alpha_data$incomplete_block)]
# Add error
alpha_data$yield <- alpha_data$yield + rnorm(nrow(alpha_data), mean = 0, sd = 350)
head(alpha_data, 10) genotype rep incomplete_block yield
1 G1 1 1.1 6690.737
2 G2 1 1.1 6256.748
3 G3 1 1.1 6364.876
4 G4 1 1.1 6398.428
5 G5 1 1.1 6426.309
6 G6 1 1.2 5131.415
7 G7 1 1.2 7174.288
8 G8 1 1.2 6985.033
9 G9 1 1.2 6968.603
10 G10 1 1.2 4985.144Model Fitting
# Fit Alpha-Lattice Model
alpha_model_fixed <- mandala(
fixed = yield ~ genotype,
random = ~ rep + rep:incomplete_block,
data = alpha_data
)
summary(alpha_model_fixed)
var_comps_alpha <- summary(alpha_model_fixed)$varcomp
print(var_comps_alpha)
# Heritability Estimation
alpha_model_random <- mandala(
fixed = yield ~ 1,
random = ~ genotype + rep + rep:incomplete_block,
data = alpha_data
)
h2_alpha <- h2_estimates(
random_mod = alpha_model_random,
fixed_mod = alpha_model_fixed,
genotype = "genotype"
)
print(h2_alpha)
# BLUPs
blups_alpha <- mandala_predict(alpha_model_random, classify_term = "genotype")
head(blups_alpha)Dataset 3: Multi-Environment Trial
Data Description
A multi-environment trial with:
- 30 genotypes
- 4 locations (environments)
- 3 blocks per location
set.seed(101)
n_genotypes <- 30
n_locations <- 4
n_blocks <- 3
met_data <- expand.grid(
genotype = factor(paste0("G", 1:n_genotypes)),
location = factor(paste0("Loc", 1:n_locations)),
block = factor(paste0("B", 1:n_blocks))
)
# Simulate yield data
met_data$yield <- 5500
# Add genotype main effects
genotype_effects <- rnorm(n_genotypes, mean = 0, sd = 300)
met_data$yield <- met_data$yield +
genotype_effects[as.numeric(met_data$genotype)]
# Add location effects
location_effects <- c(800, 200, -300, -700)
met_data$yield <- met_data$yield +
location_effects[as.numeric(met_data$location)]
# Add genotype x location interaction
gxe_effects <- matrix(
rnorm(n_genotypes * n_locations, mean = 0, sd = 250),
nrow = n_genotypes,
ncol = n_locations
)
met_data$yield <- met_data$yield +
gxe_effects[cbind(as.numeric(met_data$genotype),
as.numeric(met_data$location))]
# Add block within location effects
n_block_loc <- n_locations * n_blocks
block_loc_effects <- rnorm(n_block_loc, mean = 0, sd = 150)
met_data$block_loc <- factor(paste0(met_data$location, "_", met_data$block))
met_data$yield <- met_data$yield +
block_loc_effects[as.numeric(met_data$block_loc)]
# Add error
met_data$yield <- met_data$yield + rnorm(nrow(met_data), mean = 0, sd = 400)
head(met_data, 10) genotype location block yield block_loc
1 G1 Loc1 B1 5731.465 Loc1_B1
2 G2 Loc1 B1 5523.519 Loc1_B1
3 G3 Loc1 B1 5900.409 Loc1_B1
4 G4 Loc1 B1 6928.250 Loc1_B1
5 G5 Loc1 B1 5208.988 Loc1_B1
6 G6 Loc1 B1 5877.214 Loc1_B1
7 G7 Loc1 B1 6453.261 Loc1_B1
8 G8 Loc1 B1 5900.387 Loc1_B1
9 G9 Loc1 B1 6430.253 Loc1_B1
10 G10 Loc1 B1 6269.523 Loc1_B1Exploratory Analysis
# Location summary
met_data %>%
group_by(location) %>%
summarise(
mean_yield = mean(yield),
sd_yield = sd(yield),
min_yield = min(yield),
max_yield = max(yield)
)# A tibble: 4 × 5
location mean_yield sd_yield min_yield max_yield
<fct> <dbl> <dbl> <dbl> <dbl>
1 Loc1 6156. 488. 4924. 7290.
2 Loc2 5567. 605. 4126. 6960.
3 Loc3 5304. 536. 4058. 6528.
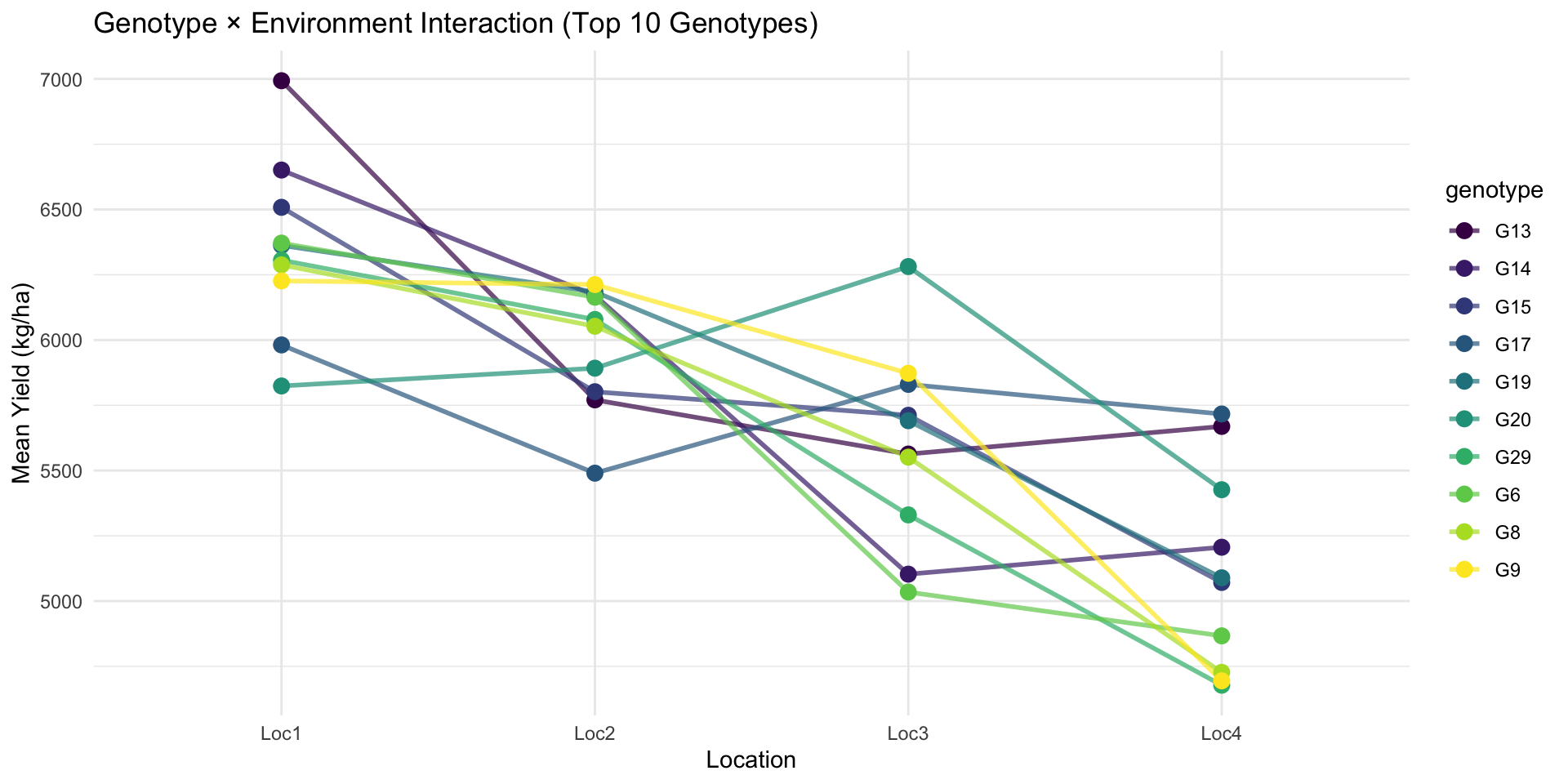
4 Loc4 4686. 545. 3584. 6042.# GxE interaction plot
met_means <- met_data %>%
group_by(genotype, location) %>%
summarise(mean_yield = mean(yield), .groups = "drop")
# Select top 10 genotypes for visualization
top_genotypes <- met_means %>%
group_by(genotype) %>%
summarise(overall_mean = mean(mean_yield), .groups = "drop") %>%
arrange(desc(overall_mean)) %>%
head(10) %>%
pull(genotype)
met_means %>%
filter(genotype %in% top_genotypes) %>%
ggplot(aes(x = location, y = mean_yield, group = genotype, color = genotype)) +
geom_line(linewidth = 1, alpha = 0.7) +
geom_point(size = 3) +
theme_minimal() +
scale_color_viridis_d() +
labs(
title = "Genotype × Environment Interaction (Top 10 Genotypes)",
x = "Location",
y = "Mean Yield (kg/ha)"
)
Model Fitting
# Fit Multi-Environment Model
met_model_fixed <- mandala(
fixed = yield ~ genotype + location + genotype:location,
random = ~ location:block,
data = met_data
)
summary(met_model_fixed)
var_comps_met <- summary(met_model_fixed)$varcomp
print(var_comps_met)
# Heritability Estimation
met_model_random <- mandala(
fixed = yield ~ location,
random = ~ genotype + genotype:location + location:block,
data = met_data
)
h2_met <- h2_estimates(
random_mod = met_model_random,
fixed_mod = met_model_fixed,
genotype = "genotype"
)
print(h2_met)
# BLUPs
blups_by_loc <- mandala_predict(met_model_random, classify_term = "genotype:location")
blups_overall <- mandala_predict(met_model_random, classify_term = "genotype")
head(blups_overall)Summary
This tutorial demonstrated:
- RCBD Analysis — Simple randomized complete block design
- Alpha-Lattice Design — Incomplete block designs
- Multi-Environment Trials — Analyzing GxE interactions
Next Steps
Session Information
sessionInfo()R version 4.5.2 (2025-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Tahoe 26.1
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidyr_1.3.1 ggplot2_4.0.0 dplyr_1.1.4
loaded via a namespace (and not attached):
[1] vctrs_0.6.5 cli_3.6.5 knitr_1.50 rlang_1.1.6
[5] xfun_0.54 purrr_1.1.0 generics_0.1.4 S7_0.2.0
[9] jsonlite_2.0.0 labeling_0.4.3 glue_1.8.0 htmltools_0.5.8.1
[13] scales_1.4.0 rmarkdown_2.30 grid_4.5.2 evaluate_1.0.5
[17] tibble_3.3.0 fastmap_1.2.0 yaml_2.3.10 lifecycle_1.0.4
[21] compiler_4.5.2 RColorBrewer_1.1-3 htmlwidgets_1.6.4 pkgconfig_2.0.3
[25] farver_2.1.2 digest_0.6.37 viridisLite_0.4.2 R6_2.6.1
[29] utf8_1.2.6 tidyselect_1.2.1 pillar_1.11.1 magrittr_2.0.4
[33] withr_3.0.2 tools_4.5.2 gtable_0.3.6